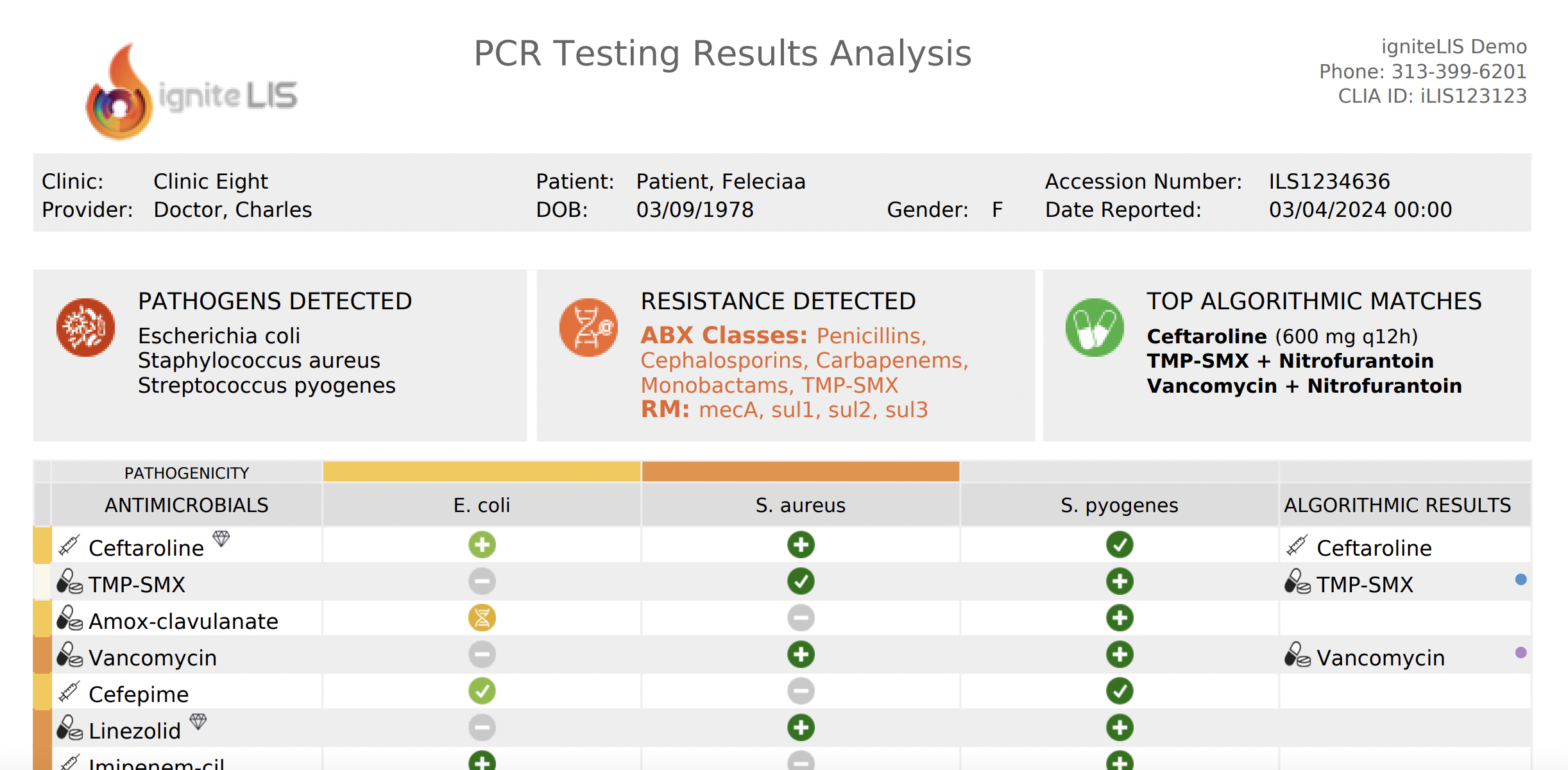
A data-driven approach to antibiotic selection.
Contact Us
We welcome any questions you may have and deeply value your feedback. It provides us with invaluable insights into how you're using igniteABX and how we can improve it to better meet your needs. Please don't hesitate to reach out!
NOTE: igniteABX reports are intended for use by licensed healthcare professionals. Patients should consult their provider for interpretation or treatment decisions.
igniteABX: explained
Click the info dots to navigate between features of the igniteABX report
FAQ
Simple answers to some of the questions you may have...
How does igniteABX compare to using an antibiogram or other resources?
While medical guides provide general treatment guidelines, by integrating additional layers of information, igniteABX arrives at a more precise assessment of susceptibility. Moreover, in scenarios involving multiple pathogens, resistances, resistance determinants, local susceptibility rates, orphan markers, and other factors, the complexity pushes the limits of efficient manual interpretation and is best suited for computer-based analysis. This is where the algorithm demonstrates its true value, providing a comprehensive and efficient method for analyzing PCR results.
Can I trust that one of the algorithmic choices is the absolute best treatment for the infection detected?
In short, no. While the report employs advanced analytics to maximize prediction accuracy, it does so within the context of inherently unpredictable biological systems and sometimes scarce scientific data. Certain limitations of PCR testing also play a role. Detected resistance markers may or may not be associated with the pathogen responsible for the disease. The presence of a resistance marker does not invariably correlate with phenotypic resistance, nor does its absence ensure susceptibility. Detected organisms could be colonizers, contaminants, or normal flora, while other, non-tested organisms might be the actual cause of infection. Additionally, the algorithm cannot account for patient-specific factors such as prior antibiotic use or clinical presentation. Despite these limitations, PCR testing can reveal unparalleled insights into an infection, and the report may be an optimal tool to interpret the results efficiently and provide insightful analysis for data-driven decision-making.
Does the algorithm take into account local resistance rates?
Yes. igniteABX algorithm calculates an "estimated susceptibility" score that is based on local resistance data and PCR testing results. Where available, it utilizes state-specific resistance data. Otherwise, it relies on regional resistance data from the NIH. As resistance rates can vary significantly from state to state or even from one facility to another, the user has the option of uploading their preferred local susceptibility data for algorithm integration, further improving its accuracy.
How are testing results and local resistance data being used to calculate "estimated susceptibility”?
When your PCR testing yields negative results, the algorithm calculates "estimated susceptibility" by adjusting local susceptibility rates for the corresponding antibiotics. This adjustment is based on the percentage of resistance attributed to the tested markers. It is worth noting that the same markers collectively contribute varying percentages in different organisms, as every species has distinct resistance determinants. No adjustment is made if a detected organism does not express these markers.
What are orphan markers?
Orphan markers, identified through PCR testing, are resistance markers not typically associated with the DNA of any detected pathogens. In our algorithm, antibiotic classes are not excluded if resistance is linked to orphan markers. Consequently, a specific antibiotic may appear in both the "Resistant To" and "Results" columns. Despite possible confusion, we highlight resistance due to orphan markers as it provides valuable insights, such as indicating the potential presence of other un-targeted organisms. In the upcoming update of igniteABX, orphan markers will be correlated with likely pathogen hosts.
What other useful features are in the works?
Additional features are planned for release, notably a report portal tailored for clinicians. This portal will offer additional information specific to each report, such as pathogen and resistance statistics, references and medical literature, infection management best practices, and comprehensive prescription guidelines with dosing calculators for medications like vancomycin and aminoglycosides. Please let us know if there's a particular feature you would like to see included.
How long does it take for the ABX report to be generated?
The ABX report is generated as soon as the PCR results are ready, allowing you to review both without any delay.
I still have questions...
We welcome any questions or comments about the igniteABX report. Through your feedback, you contribute to the evolution of our end product, helping us refine and enhance its functionality, usability, and overall effectiveness. We thank you in advance!
Can you spare a moment?
Your time and insights are immensely valued. Thank you sincerely for your participation!
Your responses are anonymous, but you can provide your name and contact information in the comments if you wish.
Did you know?
A short history
The first true antibiotic, penicillin, was discovered by Alexander Fleming in 1928. However, it wasn't until the 1940s that penicillin became widely available for clinical use. The earliest documented cases of antibiotic resistance occurred shortly after the introduction of antibiotics into clinical practice. For example, penicillin-resistant strains of Staphylococcus aureus were reported as early as the 1940s.
Deadly toll
Antibiotic-resistant infections are responsible for at least 700,000 deaths globally each year, and if left unchecked, this number could rise to 10 million deaths annually by 2050, surpassing cancer as a leading cause of death.
Quorum Sensing
Bacteria can communicate and coordinate their activities through a process called quorum sensing. By releasing and detecting chemical signals, bacteria can sense their population density and adjust their behavior accordingly, such as forming biofilms or producing virulence factors.
Economic Burden
Antibiotic resistance imposes a significant economic burden on healthcare systems and economies worldwide. In the United States, the annual economic cost of antibiotic-resistant infections is estimated to exceed $20 billion in direct healthcare costs and $35 billion in lost productivity due to illness and premature death. By 2050, the cumulative economic cost of antibiotic resistance could reach $100 trillion globally, surpassing the current global GDP.
UTI's
In the United States alone, it's estimated that UTIs account for over 8 million doctor visits annually. This prevalence underscores the significant burden that UTIs impose on healthcare systems and individuals alike.
One for the Spelling bee
Elizabethkingia anophelis is a Gram-negative bacterium that was first described in 2011. It has been implicated in outbreaks of bloodstream infections, particularly in healthcare settings. E. anophelis infections can be difficult to treat due to its resistance to many antibiotics.