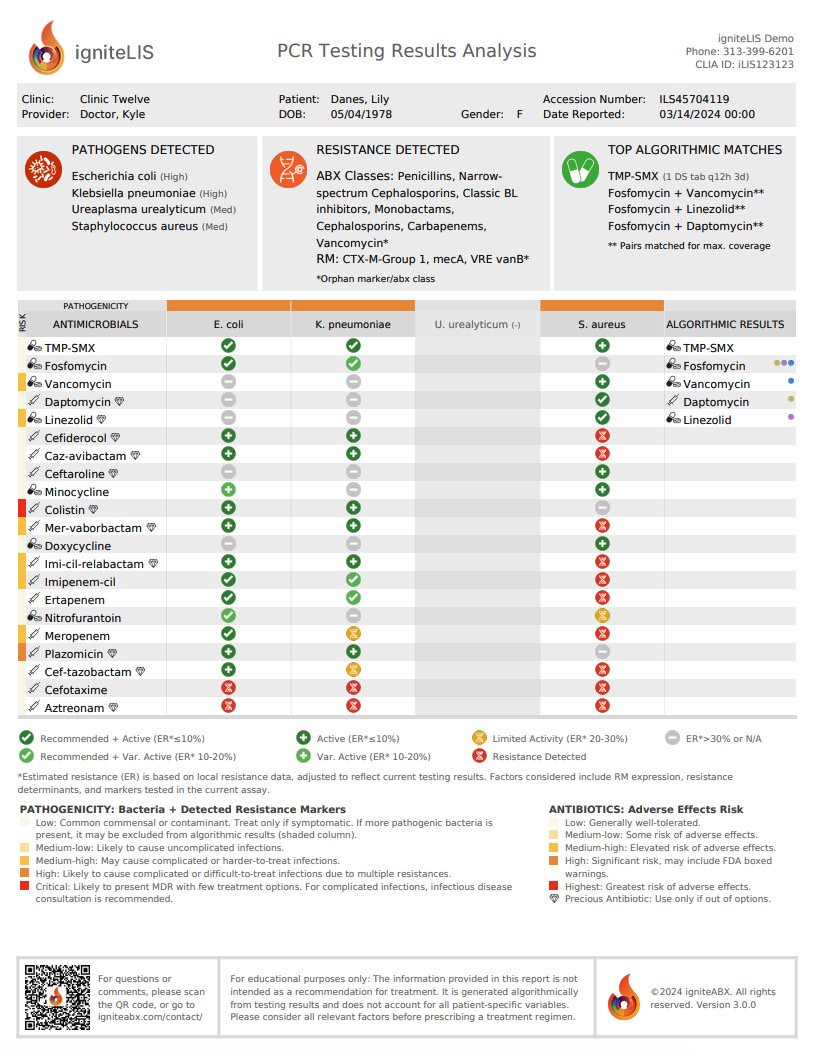
Does the algorithm take into account local resistance rates?
Yes. igniteABX algorithm calculates an "estimated susceptibility" score that is based on local resistance data and your testing results. Where available, it utilizes state-specific resistance data. Otherwise, it relies on regional resistance data from the NIH. The user also has the option of uploading their own susceptibility data.
Do I no longer need to consult my local antibiogram?
As resistance rates can vary significantly from state to state or even from one facility to another, if you have a preferred antibiogram, we encourage you to upload it to igniteABX. This ensures that the calculated "estimated susceptibility" value aligns accurately with your local data. Please reach out if you require any assistance.
How are testing results and local resistance data being used to calculate "estimated susceptibility"?
When your PCR testing yields negative results, the algorithm calculates "estimated susceptibility" by adjusting local susceptibility rates for the corresponding antibiotics. This adjustment is based on the percentage of resistance attributed to the tested markers. It's worth noting that the same markers collectively contribute varying percentages in different organisms, as each bacterium possesses distinct resistance determinants. No adjustment is made if a detected organism does not express these markers.
Can I trust that one of the algorithmic choices is the best treatment for my patient?
In short, no. Biological systems are inherently unpredictable, and pathogen-specific data is often scarce. Additionally, the algorithm doesn't consider crucial patient-specific factors, such as previous antibiotic use, clinical presentation, etc. It's important to understand that this algorithm is just one tool in your medical arsenal aimed at assisting you in making more informed decisions, but it's not a substitute for medical judgment. Please consider all relevant factors before prescribing a treatment regimen.
What are orphan markers?
Orphan markers, identified through PCR testing, are resistance markers not associated with the DNA of any detected pathogens. In our algorithm, antibiotic classes aren't excluded if resistance is linked to orphan markers. This may result in a specific antibiotic being listed in both the "Resistant To" and "Results" columns. Despite possible confusion, we highlight resistance due to orphan markers as it provides valuable insights, such as indicating the potential presence of other untargeted organisms. In the next version of igniteABX, you'll be able to see the likely pathogens hosting the orphan markers.
What other useful features are in the works?
Further enhancements are planned for release this summer, notably a report portal tailored for providers. This portal will offer additional information specific to each report, such as pathogen and resistance statistics, references and medical literature, infection management best practices, and comprehensive prescription guidelines with dosing calculators for medications like vancomycin and aminoglycosides.
I still have questions...
We welcome any questions or comments about the igniteABX report. Through your feedback, you contribute to the evolution of our end product, helping us refine and enhance its functionality, usability, and overall effectiveness. We thank you in advance!